SynPharm: A Guide to PHARMACOLOGY Database Tool for Designing Drug Control into Engineered Proteins
This paper is an account of the project I worked on from September 2015 to 2016, in Professor Jamie Davies’ lab - my first major project. I created a tool called SynPharm, which aims to solve a specific problem in the field of Synthetic Biology - so to understand the tool we first have to step back and look at what Synthetic Biology is and how it is done.
Synthetic Biology
Synthetic Biology is one of the most exciting fields in Science today. Put most dramatically, it is nothing less than humanity’s attempt to design and create artificial and new living organisms, in a laboratory, to solve the many (many) problems facing our own species in 2018. We can create bacteria which could clean up oil spills, synthesise materials for us, and ultimately (some day) even modify our own bodies. The potential, and the challenges, of such an endeavour are both enormous.
While there are some groups who really are attempting to design new living systems entirely from scratch, in practice Synthetic Biology usually involves inserting new genes into existing organisms, to give them new abilities. Genes are stretches of DNA which contain instructions for making a protein, and it is proteins which do almost everything useful in life - give an organism a new gene, and it will gain all of the abilities of the resultant protein. Want a bacterium to gain the ability to suck up copper from the environment? Insert into it a gene which provides instructions for making a protein which does just that.
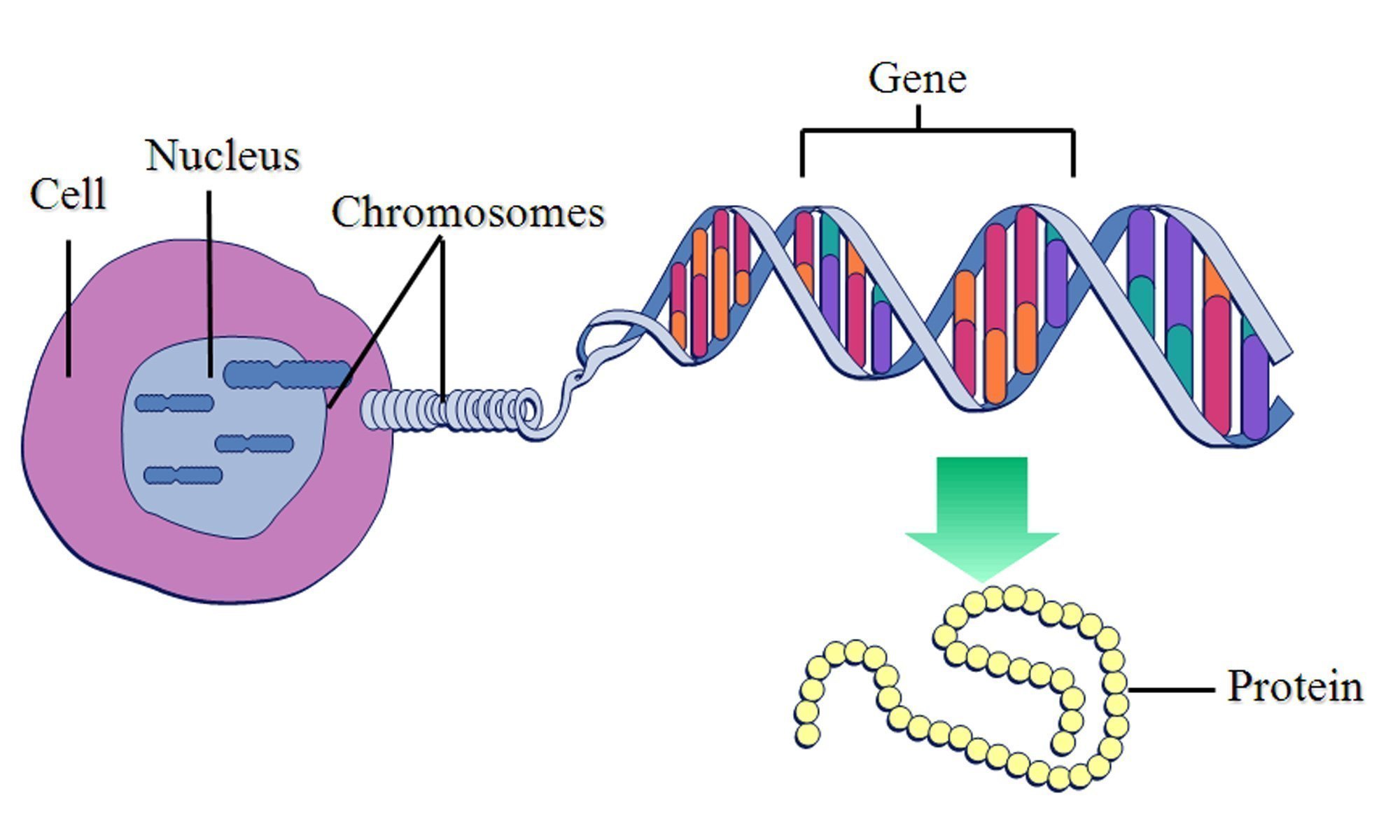
A diagram I stole from this study illustrating this principle.
But it is more than just putting in a gene (or genes) and hoping for the best - Synthetic Biology above all emphasises its engineering credentials. It focuses on modular, reusable parts that can be inserted into an organism of choice with little to no knock on effects to the rest of the host organism's genetic systems, and which have predictable, host-independent properties. That’s the theory anyway - the realities of Biology's chaotic workings often make this difficult in practice.
So, in the 'traditional approach' to Synthetic Biology (if there can be such a thing for such a new discipline), you introduce a new gene into an organism - a bacterium, a plant, or even an animal - and it can now produce a new protein that does something useful, such as make a chemical, or break down a harmful toxin. Since you probably don't want this protein to be produced all the time, you will need a way to turn on or off the production of the protein. Usually this is done by making the gene dependent on some small molecule - when you apply a solution of this molecule, the gene will either turn on and start making the protein, or turn off and stop making the protein.
This traditional approach has two problems however. The first is that controlling the new protein by acting on its gene is a bit 'laggy'. There is a not-unimportant delay between the gene being turned off, and the protein actually disappearing - as there will still be protein already made in the cell and that takes time to break down. Likewise turning on a gene will require time before production can be ramped up. This unresponsiveness to external control can be at best inconvenent, and at worst deadly - depending on the application.
A second problem occurs when such synthetic genes are introduced into humans - not something that generally happens now, but which is on the horizon due to the tremendous health benefits that cound ensue if done right. The small molecule used to control the turning on and off of the genetic system would have to be given to the person - as an injection or as a pill or some other vehicle - which means it will need approval from the FDA or some other body to ensure it’s safe. This takes, to put it mildly, an enormous amount of time and money before your new genetic system can be put into operation.
Which brings me to my project, and the approach suggested to solve both of these problems.
Inverse Pharmacology
The proposed solution to both of these problems is what Professor Jamie Davies, my supervisor for this project, has called 'Inverse Pharmacology'.
The idea is that you can solve the problem of delay in the system by still using a small molecule to turn on or off the system, but instead of targeting the gene you instead target the protein product - activating or inactivating it immediately when the control substance is added. This is less straightforward that it might at first seem - genes have built in machinery called transcription factors for allowing their activation to be controlled by signals, which is why most synthetic gene systems use such control - protein molecules have no such built in responsiveness.
But, many proteins do respond to small molecules anyway, and change their activity in response. In fact there is an entire field devoted to taking advntage of this - pharmacology. Traditionally pharmacology works by identifying an existing, natural protein target that you want to modify, then searching for a small molecule that will bind to it - and then making it into a drug. The idea behind inverse pharmacology is that you instead search through known drug targets for those whose drug responsive nature can be transfered to another protein.
How does that work? Well, evolution - the process which sculpted all these proteins in the first place - rarely creates proteins entirely from scratch. Instead it combines existing functional units and swaps their orders and sequence, resulting in many proteins that are organised into distinct, self-contained functional ‘domains’. If a drug target has a small, discrete drug responsive domain that is distinct from the domain that carries out the restof its function, that domain can be transfered to another protein.
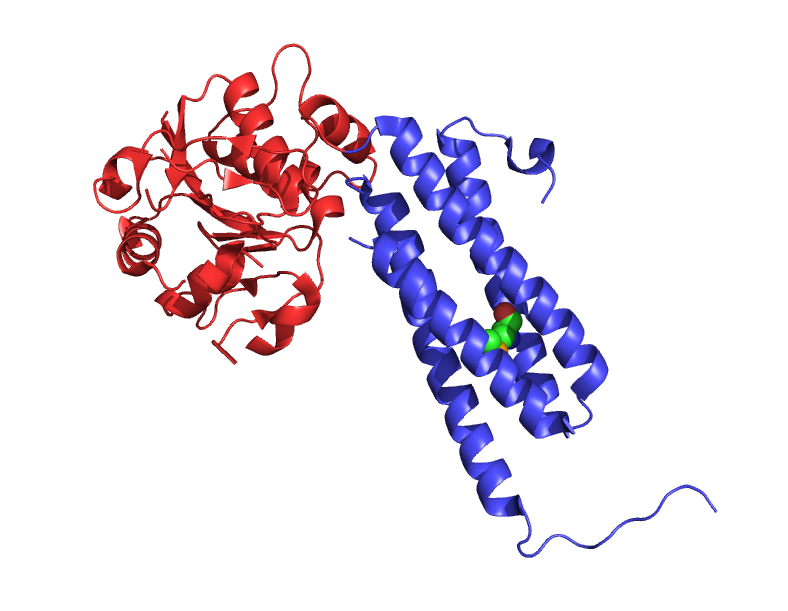
A protein with two protein domains, shown in different colours. They are a single molecule, connected to each other, but fold and act independently. One domain can be seen binding to a smaller molecule.
So, you could create a synthetic protein which does... whatever it is you want it to do, but also has a domain added which responds to a small molecule. Then create the gene instructions for this new protein and insert it.
The beauty of this system is that because you are using protein domains that respond to pharmacological drugs - in many cases the small molecules you would use to control your system already have FDA approval - thus also at a stroke solving the second problem.
So, what Jamie was proposing was that Synthetic Biology could turn its attention to gene systems where the protein product itself (rather than the gene) responds to your control signal, and that this should be brought about by adding drug responsive protein domains to them, which respond to safe-for-humans small molecules.
SynPharm
So, in September 2015 I was hired by Jamie to create a software tool for Synthetic Biologists that would allow them to select and design such synthetic proteins. One of the reasons Jamie’s lab is well placed to deliver this - other than their many existing contributions to (especially Mammalian) Synthetic Biology - is that attached to the lab is the Guide to Pharmacology team.
The Guide to Pharmacology (GtoP), as I have written about previously, is an expert-curated database of drug targets and the molecules that act on them. It contains information on interaction affinity, approval status, and much else besides, and the data is accessible over the web, and via a web API - the perfect resource to start with.
The Guide's homepage as of January 2018.
So, over the course of 12 months I built SynPharm, and the pipeline needed to populate it. I started by writing scripts which went through all interactions in the GtoP database (15,000 or so at that time) and looked for those for which there existed structural data - that is there are atomic structures from experiments of the drug actually bound to its target. After some filtering, my pipeline then identifies the binding residues, and tries to work out which portion of the target protein mediates drug binding. This information is then saved to a separate database, and is accessible via the web interface that I built at the same time.
Using SynPharm then, a researcher can see the drug responsive elements of (to date) about 600 drug targets. In each case they can see estimates of its suitability to function independently, information on the drug which binds to it, and links back to the GtoP for more indepth information. There are also 3D views of the drug binding, and colourful representations of internal contacts. They can, it is hoped, then select a suitable module for inclusion in their synthetic gene systems that would be responsive and safe.
(The paper itself gives a more detailed account of what the website provides and how the data is generated.)
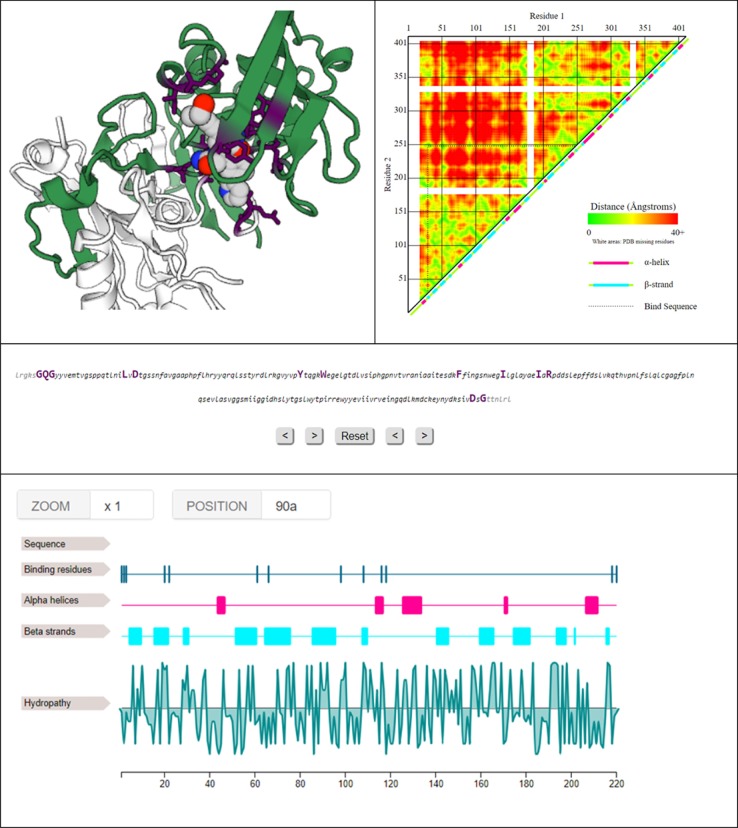
A summary of some of the information SynPharm provides.
I very much enjoyed this project, and am proud of the end result. I was fortunate to work on this within the team that I did - as already mentioned Jamie’s lab was well placed to work towards this, having such highly skilled people within both Synthetic Biology and Pharmacology. I received invaluable help from both of these groups, and am very grateful for this - as my first post-graduation project this could have been very daunting and difficult to begin with, but I could not have asked for a better welcome or more useful assistance from the rest of Jamie’s lab.
The group continues to work on Inverse Pharmacology and its applications, and I hope SynPharm can be a useful component in these endeavours.